It’s Clear as Day: HIF Signaling is Driving Force of the Clear Cell Morphology
Whitney A. Brown, W. Kimryn Rathmell*, Zachary A. Bacigalupa* Department of Medicine, Vanderbilt University Medical Center, Nashville, TN 37232ABSTRACT
Clear cell renal carcinoma (ccRCC) is the most common form of kidney cancer with few therapeutic options in its advanced stages. ccRCC has genetic predisposition linked to the von Hippel Lindau gene. The product of this gene is responsible for proteasomal degradation of the hypoxia induced factors, which when stabilized activate hundreds of pathways, some of which promote tumor growth via angiogenesis, and upregulating glycogen and lipid biosynthesis. The “clear cell” morphology exhibits a large, translucent cytoplasm attributed to excessive glycogen and lipid deposition. Biochemical analyses have demonstrated that these lipid depots in ccRCC are enriched with high concentrations of high-density lipoprotein cholesterol, which is known to play an integral role in membrane rigidity and drug resistance. Glycogen synthesis serves as an energy source for tumoral growth, and lipid and cholesterol buildup within tumors has been linked to the formation of new cell membranes for cellular growth. In this review we will summarize how glycogen, lipid, and cholesterol metabolism play key roles in ccRCC tumor growth and the therapeutic potential of targeting these pathways.
INTRODUCTION
Clear cell renal cell carcinoma (ccRCC) is the most common form of kidney cancer, accounting for 70-75% of all kidney cancers, which affects males twice as often as females ¹. Current therapies include tyrosine kinase inhibitors (TKI) targeting factors involved in angiogenesis, which is essential for ccRCC tumor growth², ³, immunotherapies, targeting checkpoints regulating T cell activation4, and the combination of both5. Identifying strategies to enhance the efficacy of current therapeutics, or to achieve durable disease control with reduced toxicity, has become the focus of current investigations. ccRCC is linked to genetic factors that control cell metabolism, which makes it a ripe target for studying the oncologic metabolic shift known as the Warburg effect5 as a potential therapeutic angle. The Warburg effect describes a dependence on aerobic glycolysis and lactic acid fermentation, while the tricarboxylic acid (TCA) cycle is downregulated even in the presence of oxygen. Studies have shown an increase in glucose uptake and aerobic glycolysis 6-9. Fewer TCA intermediates were present in ccRCC, further confirming a shift towards aerobic glycolysis and indicating that pyruvate dehydrogenase is less active in ccRCC6,10. This discovery also demonstrates that ATP production is dependent on aerobic glycolysis rather than oxidative phosphorylation6,10. Within the TCA cycle, fumarate and malate levels were lower than normal tissues, while succinate, isocitrate, and citrate were higher, indicating a dependence on reductive carboxylation through citrate8,9. This upregulation of reductive carboxylation was shown to be the route for fatty acid synthesis in ccRCC11-13. Given that a Warburg shift is a complex matter with many intermediates, this discovery in ccRCC provides multiple targets for therapeutic interventions; currently glutaminase inhibitors are being examined as target to prevent the formation of citrate, and therefore prevent reductive carboxylation in ccRCC13.
These genetic predispositions in ccRCC are linked to chromosome 3 translocations, deletions, and mutations that effect the von Hippel Lindau (VHL) gene and its expression. This molecule is well known as a major effector of the hypoxia response, as the key negative regulator of the hypoxia inducible factors (HIF), a potent family of transcription factors and their downstream transcriptional targets such as vascular endothelial growth factor (VEGF)14–16. HIFs interact with the product of VHL (pVHL) through oxygen dependent domains that are targeted prolylhydroxylation enzymes 15,17–19. Under normal oxygen conditions, pVHL forms a ubiquitin ligase complex that recognizes hydroxylated proline residues and binds to the alpha subunit of HIF, leading to its polyubiquitination and degradation16. In hypoxic conditions HIF-α is not recognized by pVHL, allowing it to dimerize with HIF-ß. This dimer is an essential transcriptional regulator of hundreds of genes and signaling cascades that promote hypoxic adaptation16, such as the activation of vascular endothelial growth factor receptor (VEGFR) signaling20. The HIF transcriptional network activates many enzymes and proteins integral to key metabolic pathways whose enhanced activity promotes tumor growth when pVHL is absent21, 22.
The alpha subunit of HIF is present in two main forms—HIF-1α and HIF-2α. These both have different functions in the cell and presentation in ccRCC, and this distinction is critical for discussions of metabolism. Although both HIF factors are targets of pVHL, HIF-1α is not always present in ccRCC, and VHL-mutated tumors can be classified as expressing both HIF-1 and HIF-2 (HIH2), or HIF-2 only (H2)23. The downregulation of HIF-1α is one feature that drives more aggressive disease states16 and suggests that HIF-1α has tumor suppressor functionality in ccRCC. While HIF-1α expression and activity cannot completely counteract the oncogenic effects of HIF-2α, its presence can decrease the severity of the prognosis16. When stabilized, HIF-1α, as a transcription factor, has potent effects on genes involved in activating aerobic glycolysis24,25. HIF-2α is expressed in all VHL-/- ccRCC and its elimination in these cells prevents tumor growth. The role of HIF-2α inhibition is to block HIF-2α transcription and therefore inhibit its downstream targets, such as VEGF, as well26. Studies have shown decreased tumor formation in xenograft models when HIF-2α is inhibited and pVHL is absent27-29. An effective mechanism of inhibition has been identified as inhibiting translation of HIF-2α by targeting the binding of its iron responsive element (IRE)27, 30–32. This study showed that hypoxia increases HIF concentration via a 5’-UTR IRE that binds to iron responsive protein 1 (IRP1), and when exogenous iron is added, translation of HIF proteins increases30, 33*. Additionally, a recent study showed via proximity ligation assays that an inhibitor of HIF-2α, PT2385, decreased HIF-2α complexes in ccRCC biopsies analyzed before and during treatment3434. In this study, they measured efficacy based on three factors: (1) the concentration of a downstream target of HIF-2α, erythropoietin (EPO), (2) the dissociation of HIF-2 complexes, and (3) the amount of gene expression. They found significantly decreased levels of EPO in 90% of patients after two weeks, showing the HIF inhibition was effective34. Using fluorescently conjugated antibodies for HIF-2α and HIF-1β, they were able to detect proximity via florescence microscopy to show a significant decrease in HIF-2α complexes during drug treatment as compared to pretreatment observations in two of three patient samples, and via RNA-seq analysis they found that 277 genes were downregulated by the inhibitor in those same two patients34. Complex dissociation and gene expression were found to be correlated to one another, indicating that downregulation of HIF-2α dependent genes may be necessary for antitumor activity34. Since this inhibitor was shown to have high variability, it was later improved to PT2977 and is now known as MK6482. The improvements were made with the goal of improving pharmacokinetic stability by decreasing binding to serum proteins, increasing the binding affinity for the HIF-2α binding pocket, and lowering the susceptibility of glucuronidation to a key hydroxyl group26, 35–37. A phase I trial with MK6482 concluded that 67% of patients had reduced target-lesion size with manageable anemia being the most common adverse event, and hypoxia being the only adverse event that caused patient discontinuation/dosage reduction26, 38, 39. A phase II trial used a cohort of patients with VHL-associated, nonmetastatic ccRCC; 87% of the cohort had decreased tumor size26, 40. A phase III trial is currently being conducted to compare the efficacy of MK6482 versus everolimus26, 41. The mechanism of resistance to HIF-2α inhibitors has been identified as either mutations that prevent drug binding or mutations that increase HIF stabilization26, 34, 42, but newer HIF-2α inhibitors have the potential to overcome these mutation barriers by using a combinatorial approach, targeting factors that are implicated when resistance occurs26, 43–47. Inhibitors of HIF-2α show great clinical promise alongside other targets in ccRCC.
Another target with approved therapies for RCC treatment is the mammalian target of rapamycin (mTOR). This classical metabolism regulator is a serine/threonine kinase that functions as a nutrient sensor by responding to environmental conditions, such as changes to oxygen levels, metabolite abundance, amino acids and growth factors48. Rapamycin (sirolimus), and rapamycin analogs everolimus and temsirolimus, block mTOR activity by forming a gain-of-function complex with FK506-binding-protein (FKBP12)12–14. This complex acts as an allosteric inhibitor of mTOR complex 1 to accomplish this inhibitory effect48, 51. In addition to regulating metabolic responses, this factor acts upstream of VEGFR to further promote angiogenesis. In vitro experiments have shown that inhibition of mTOR prevents angiogenesis and tumor growth as well as decreasing lipogenesis48. We will continue to discuss specific targets within glycogen metabolism, lipid metabolism, and cholesterol metabolism for the remainder of this review.

Glycogen Metabolism
ccRCC is classified by highly regulated lipid and glycogen metabolisms and increased deposits in the cell for both52. In general, activation of glycolysis and inactivation of the TCA cycle is associated with ccRCC and explains the energy supply for the tumor53. Furthermore, there is evidence that oxidative phosphorylation is inhibited in ccRCC, which further supports that the energy supply of these tumors is dependent on glycolysis53. Specifically, high concentrations of glycolytic enzymes, which are supported by a hypoxic microenvironment, and low concentrations of TCA cycle intermediates are found in these tumor cells52. In ccRCC cells, lactate is also upregulated, in part due to transcriptional activation of Lactate Dehydrogenase (LDH), further suggesting that the cells function on aerobic glycolysis52, 54.
Although these trends are seen across the spectrum of ccRCC tumors, quantitatively, glycogen and lipid deposits are tumor grade dependent, with glycogen and lipid accumulation more prevalent in lower grade tumors54. These features have been linked to prognostic algorithms, such as the transcriptional ccA and ccB signature55, 56. Further investigations into the metabolic shifts associated with stage progression are being described with increasing frequency, most recently with the Cancer Genome Atlas index paper on ccRCC5, 7 and dedicated metabolomic profiling9. Finally, failure of antitumor therapies has also been linked to the expression of glycolytic and hypoxia factors and presumed upregulation of compensatory signaling pathways52*.
Glycolysis and glycogen synthesis are regulated by several factors in the cell. As discussed previously, mTOR promotes tumor growth and angiogenesis in ccRCC. One way mTOR accomplishes this is by activating glycolysis and glycogen synthesis, providing an energy source for the tumors. A recent study showed that the phosphoinositide 3-kinase (PI3K)-protein kinase B (AKT)- mTOR signaling axis is associated with the progression of ccRCC57. Human ccRCC cell lines CAKI-1 and RCC4 were treated with NVP/MEZ235, a dual inhibitor of both PI3K and mTOR, and showed decreased phosphorylation of AKT protein and mTOR. By effectively blocking AKT and mTOR activation, the researchers observed significant inhibition of glycolysis and glycogen synthesis, removing the energy source and decreasing tumoral growth57. As a tyrosine kinase that orchestrates a robust signaling cascade regulating many biosynthetic processes, PI3K has long been an integral target for TKI treatments58.

Another key regulator of glucose metabolism is glycogen synthase 1 (GYS1)59. Glycogen synthase is a major regulator of glycogen catabolism which, when active, promotes the synthesis of glycogen. A recent study showed that GYS1 is significantly overexpressed in ccRCC tumors and was mostly found in the cytoplasm, which is where glycogen synthesis occurs. This overexpression was then correlated to poor overall survival in the clinical setting59. Additionally, this study showed in a western blot that p65 expression increased when GYS1 was overexpressed via, indicating that GYS1 interacts with the canonical NF-κB pathway. Glycogen synthase is inactivated in the body by glucagon and epinephrine, so finding treatments that mimic these effects in tumor cells and treating in combination with inhibitors of glycolysis, could be an area for further investigation.
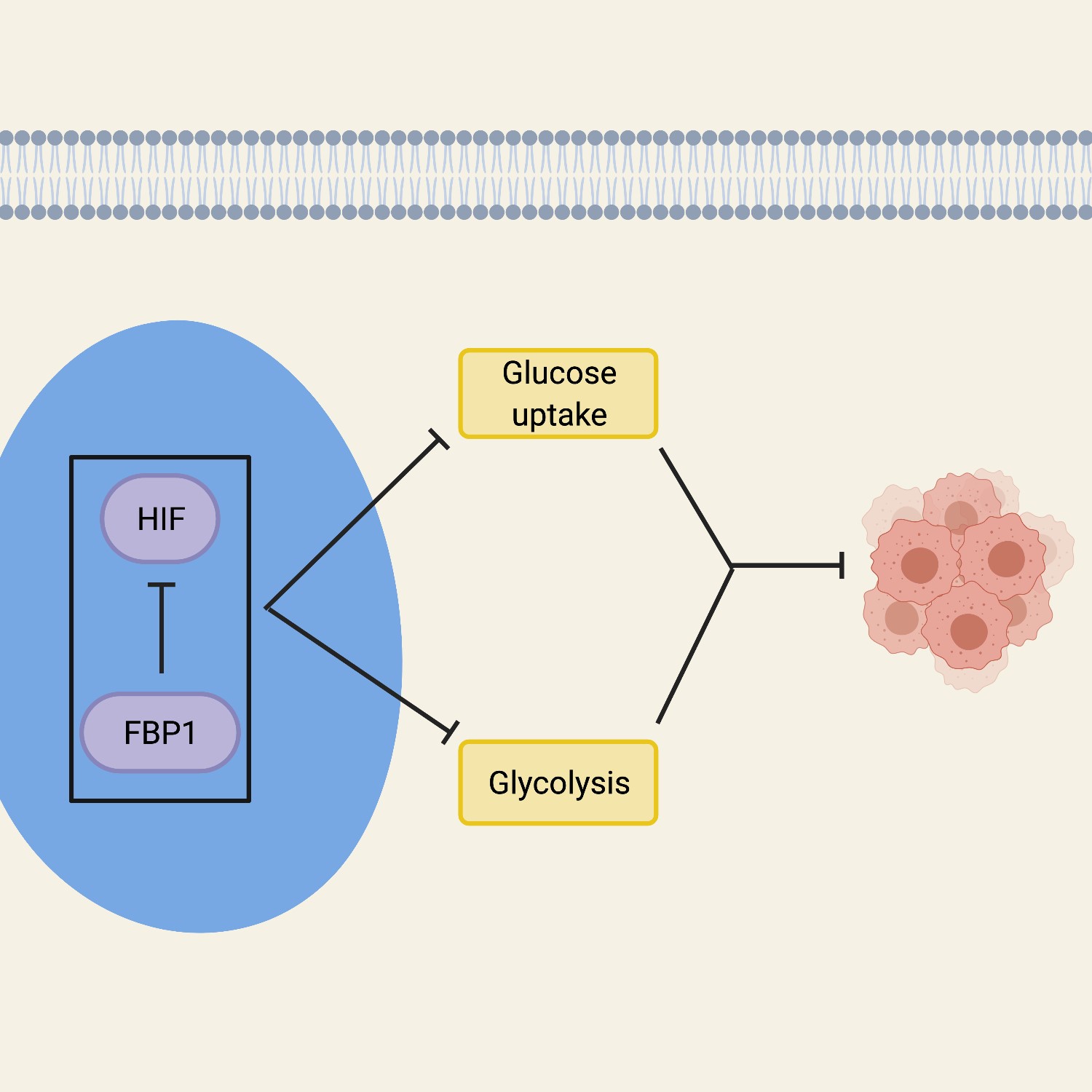
In addition to factors that promote the expression and activity of glycolytic enzymes for energy generation, several cellular modifications have been observed which suggest the regulation of this bioenergetic pathway is tightly controlled. Fructose-1,6- bisphosphatase 1 (FBP1) is a rate-limiting gluconeogenic enzyme that plays a large role in glucose metabolism and inhibits HIF proteins in the nucleus54, 60. FBP1 opposes ccRCC by inhibiting glycolysis and cell proliferation in cells52, 60. Inhibition of FBP1 increases glucose uptake and, therefore, allows tumor growth to progress. Evidence supported by cellular fractionation and immunofluorescent staining suggests that FBP1 suppresses HIF proteins in the nucleus, and showed that an interaction between FBP1 and HIF proteins is necessary for an effect on glucose metabolism60. This was further proven by using a nuclear-excluded form of FBP1 which failed to inhibit the HIF proteins in the cell, showing that the effects of FBP1 inhibition originate in the nucleus60. Overall, the FBP1 activity in the cell that affects the growth and development of tumors, works by regulating HIF from the nucleus. The inhibition of FBP1 promotes glycolytic functions, thereby enhancing the Warburg effect, while simultaneously failing to suppress nuclear HIF function, both of which is associated with poor prognosis in ccRCC (Figure 1).

Lipid Metabolism
In ccRCC, lipid metabolism is an important factor for tumor cell growth because it provides the membrane structures for the newly formed tumor cells. Specifically, lipid droplet buildup serves as fuel for membrane synthesis for these tumor cells24–26. This process of lipid droplet buildup occurs through increased lipogenesis via reductive carboxylation in parallel with the inhibition of beta-oxidation11–13, 61. Evidence shows that increased lipid storage in ccRCC cells is associated with increased tumorigenesis, and there is a correlation between lipid metabolism and ccRCC risk score62, 63. A recent study looked into the effects of VHL status on lipid catabolism versus lipid uptake. By staining with Oil red O to assess changes to the presence of lipid droplets, Du et al. observed a decrease in lipid droplets in cells where VHL was reconstituted, suggesting that the presence of pVHL impacts either lipid uptake/synthesis or promotes lipid catabolism62. In an effort to interrogate the effect on lipid uptake, this study tracked the uptake of BODIPY fluorescent fatty acid dyes and concluded that lipid uptake occurred independently from VHL status62. Therefore, lipid deposition is VHL-mediated while lipid uptake occurs independently of VHL, indicating that de novo lipid synthesis is the major contributor to lipid droplet formation in VHL-/- ccRCC62. Several factors in the cell regulate this process and are currently being studied as points of therapeutic intervention.
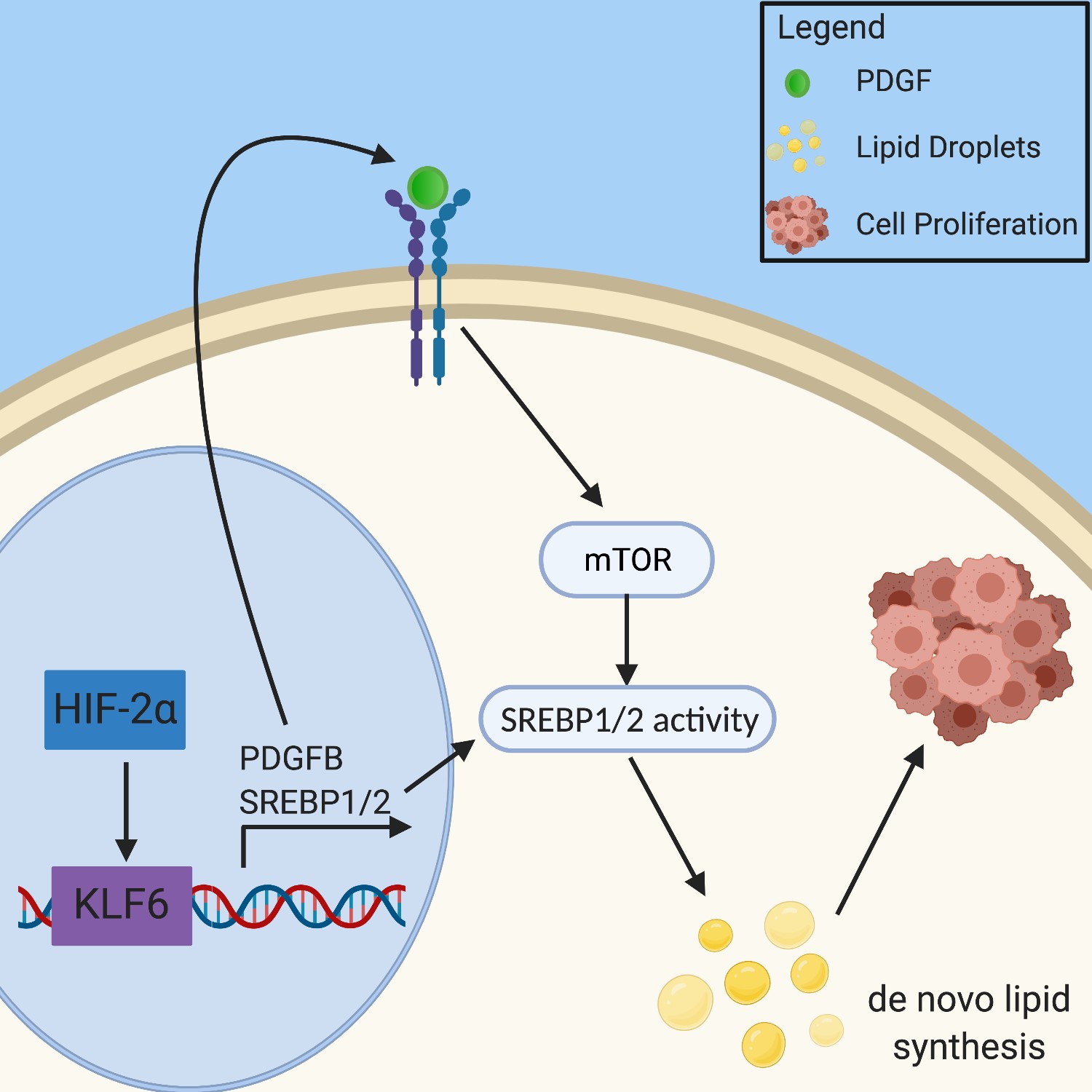
One regulator of interest is Kruppel life factor 6 (KLF6). KLF6 is a zinc finger family transcription factor that was shown to have effects on lipid metabolism64 and has been implicated as a tumor promoting factor in ccRCC via its effects on cell proliferation and high levels of expression. The gene encoding this transcription factor was found to be located within a locus containing one of the strongest super enhancers. Additionally, this association was linked to enhanced KLF6 expression when comparing ccRCC samples to adjacent normal tissue, as well as to other solid tumors lacking this super enhancer. The Cancer Genome Atlas data of ccRCC showed a correlation between HIF-2α expression and KLF6 expression; this study investigated this interaction through VHL reintroduction experiments64. The reintroduction of VHL caused a decrease in mRNA expression of KLF6 and, using ChIPseq, they showed that VHL introduction caused a decrease in activity in the region where the super enhancer is located64. Additionally, the ChIP-seq data show that HIF-2α was bound at this same region64. This indicates that HIF-2α is an activator of this super enhancer, so when HIF-2α is present, it binds to the super enhancer and there is robust transcription of KLF6. To expand on their findings, the researchers next assessed the impact of altering KLF6 expression in ccRCC. Pathway analysis was performed on RNA-seq data collected from cells depleted of KLF6 and revealed a significant downregulation of lipid and cholesterol metabolism pathways64. Specifically, they identified sterol regulatory element binding protein 1 and 2 (SREBP1 and SREBP2), master transcriptional regulators of lipid signaling, were downregulated in response to KLF6 suppression. These findings were validated with qPCR experiments, where it was observed that SREBP1, SREBP2, and several of their downstream targets were downregulated in response to KLF6 inhibition. Importantly, these results translated further into an overall decrease in intracellular cholesterol and lipids when KLF6 is depleted. These studies elegantly display the critical role HIF-2α plays in regulating KLF6, an essential piece of lipid and cholesterol metabolism in ccRCC.
mTOR signaling through mTORC1 also regulates SREBP1 and SREBP2. Investigations into the interaction between mTORC1 and KLF6 revealed that KLF6 both directly interacts with SREBP1 and SREBP2, and promotes mTOR signaling by enhancing platelet-derived growth factor subunit B (PDGFB); both of these factors contribute to an increase in lipid metabolism and anabolic signaling, resulting in increased tumor growth64 (Figure 2). SREBP acts by inducing the production of enzymes involved in cholesterol and lipid synthesis, including the rate-limiting enzyme of cholesterol synthesis, 3-hydroxy-3-methyl-glutaryl-coenzyme A reductase (HMGCR)65–67. A recent study showed that the gene TRC8 represses the translation of these key transcription factors, therefore inhibiting lipid and cholesterol synthesis, which makes it a target for future investigation65.
HIF proteins promote lipid metabolism via a variety of mechanisms. HIF proteins promote dietary lipid uptake, interact with the gene PLIN2 to promote lipid storage, and interacts the gene encoding carnitine palmitoyl transferase 1 (CPT1A) to promote lipid droplet formation. Lipid droplet formation was shown to be HIF protein dependent; cells that were double knockdown for HIF-1α and HIF-2α had a significant decrease in lipid droplet formation62. Additionally, this study showed that HIF-1α and HIF-2α bind specifically to a CPT1A promoter via ChIP analysis with HIF-1α and HIF- 2α antibodies in 12 regions identified as HIF response elements62. A recent study showed that dietary lipid uptake leading to increased lipid in the kidneys being driven by HIF-1α signaling in human ccRCC12. The gene PLIN2 was found to be over expressed in ccRCC and suggests an interaction with HIF- 2α allows for heightened lipid storage. The mechanism by which this occurs is through stabilization of the endoplasmic reticulum (ER). The interaction between PLIN2 and HIF-2α is required to maintain ER homeostasis and prevents cell death under stressful conditions68. This is a possible explanation for drug resistance; when the ER is targeted by therapeutic interventions, this interaction could be preventing apoptosis. Another study further analyze the HIF dependence of lipid droplet formation by focusing on the interaction between HIF proteins and the gene encoding CPT1A, which is a major regulator of lipid synthesis. When CPT1A was in low concentrations, it has shown increased lipid storage associated with tumorigenesis. It was discovered that HIF-1α and HIF-2α directly bind with CPT1A to inhibit its function and therefore increase lipid droplet formation62.
Another enzyme intimately involved in lipid metabolism is hydroxyacyl- CoA dehydrogenase alpha subunit (HADHA). The role of HADHA in regulating lipid droplet formation has been examined in several models of ccRCC, including the ccRCC cell line 786-O. In this cell line, OmicsNet and STRING analysis revealed an abundance of enzymes involved in lipid metabolism, including HADHA and acetyl-CoA acetyltransferase 2 (ACAT2), exist in a network. Additionally, several direct protein-protein interactions were identified in this network, including a link between HADHA and ACAT2, which allows them to interact with substrates in a coordinated manner69, 70. HADHA was shown to activate ACAT2, an enzyme directly involved in lipid breakdown, so at low HADHA levels, there are low levels of lipid breakdown causing lipid stores to be maintained, which is associated with ccRCC tumor cell proliferation69. In a separate study, it was confirmed that there is downregulation of both HADHA and ACAT2 in ccRCC patient tissues and that this downregulation of HADHA expression in ccRCC tumors was associated with better patient survival70. The goal in studying lipid metabolism of ccRCC is to identify opportunities to intervene therapeutically inhibiting the rapid proliferation and expansion of cells present in the tumor, as well as impeding formation of new cells. KLF6, PLIN2, HIF- 2α, HADHA, ACAT, and CPT1A are only a few of the lipid regulators that have been identified for discussion in this review, but the findings linked to these mediators suggest avenues that effect lipid droplet buildup could be attractive targets for metabolic factors incorporated into ccRCC prognosis and treatment.

Cholesterol Metabolism
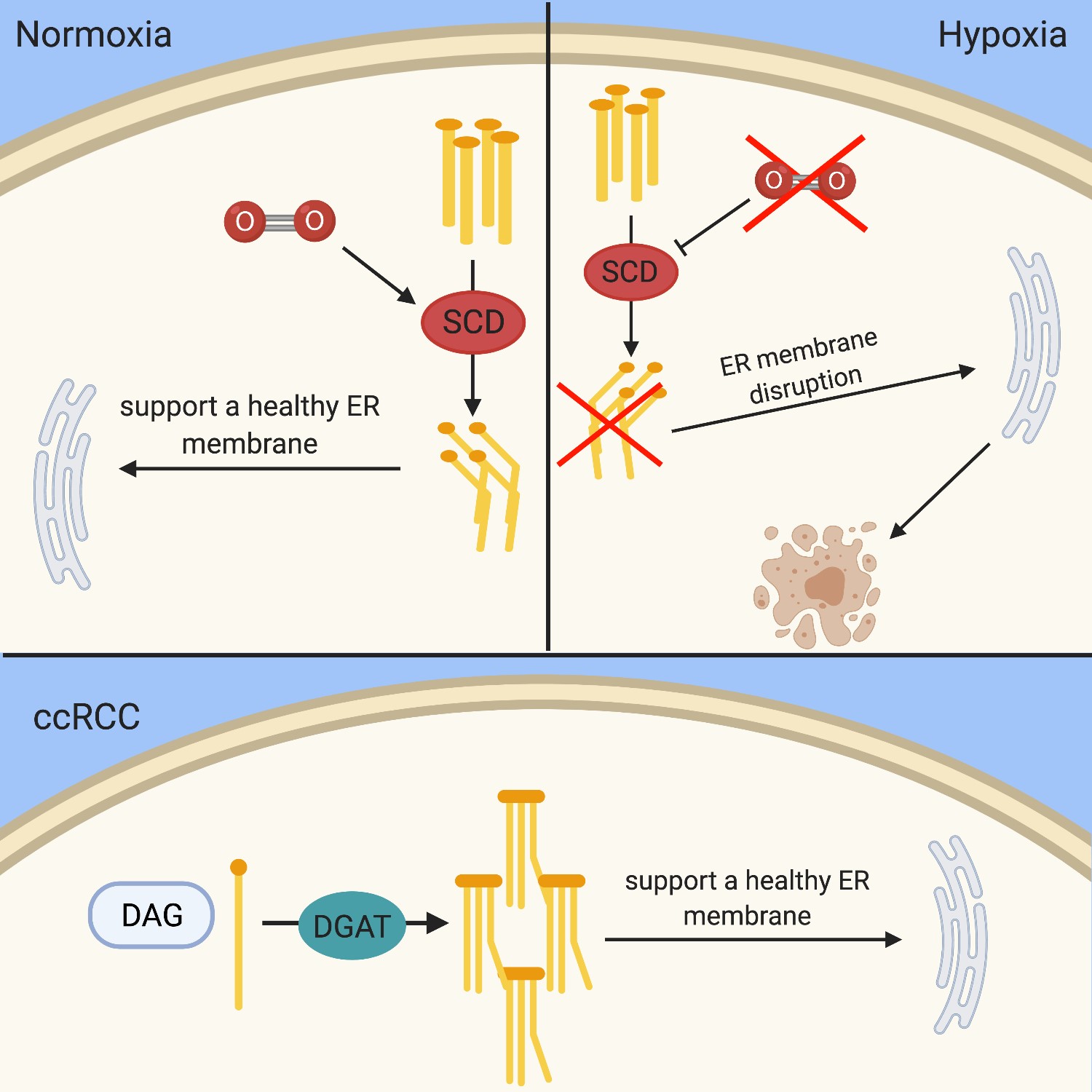
The clear cell phenotype is characterized by lipid buildup, but recent studies have shown that high-density lipoprotein (HDL) cholesterol is accumulated in the highest levels within ccRCC tissues. HDL-cholesterol is also seen in higher amount in ccRCC tumoral cells compared the surrounding non-malignant kidney tissues71–73. The deregulation of cholesterol compounds with the accumulation of other lipids to stabilize the membrane of the tumoral cells and increases tumorigenesis when it cannot be regulated properly. In multiple studies, cholesterol synthesis did not appear to be affected, which suggests that the cholesterol buildup seen within the cells is due to exogenous cholesterol influx and endogenous cholesterol efflux71, 74. Cholesterol was also discovered to play a role in promoting metastasis of ccRCC75. Hypoxia effects fatty acid saturation via the oxygen dependent enzyme stearoyl-CoA desaturase (SCD). SCD under hypoxic conditions is inhibited, which leads to a buildup of fatty acid precursors in the cell76. This leads to disruption of the endoplasmic reticulum and induces apoptosis76–78 (Figure 3).
A recent study demonstrated how cholesterol buildup in tumoral cells is due to the uptake of cholesterol rather than synthesis71. The cholesterol synthesis rate limiting enzyme HMGCR was inhibited in tumors containing higher levels of cholesterol, suggesting that cholesterol de novo synthesis is unlikely to be occurring in the tumor cell. Furthermore, they showed that the receptor for HDL-cholesterol, scavenger receptor B1 (SR-B1), which is usually in very low concentrations in the cell, had elevated levels in tumors containing high levels of cholesterol71. Another study explored the difference in predicted treatment efficacy by targeting the transcription factor receptor, liver X receptor (LXR) with an agonist versus an inverse agonist. The agonist used was LXR623 and the inverse agonist was SR9243. Both inhibited cell proliferation and induced apoptosis, but by different mechanisms. LXR623 killed tumor cells by promoting cholesterol efflux and inhibiting cholesterol influx. SR9243 upregulated the HMOX2 gene which reduced the angiogenic potential and proliferation, and it also caused a decrease in intracellular triglycerides. Neither affected the cholesterol synthesis pathway74. This makes these therapeutic targets attractive for future consideration because the synthesis of cholesterol is the main mechanism of cholesterol accumulation in normal cells. Since there is little to no new synthesis of cholesterol in ccRCC tumoral cells, but rather change in how much cholesterol is moving into the cell, the cholesterol receptors can be targets for therapeutic intervention with a potential window of specificity for tumor cells in this case.
Although high cholesterol levels
are common to all ccRCC tumors,
cholesterol levels in the body have also
been associated with outcome in the
case of ccRCC. High HDL-cholesterol
levels were correlated with better outcomes
and can act as a similar predictor
in other forms of cancer as well79. The
mechanism by which this is achieved
is believed to be that the higher HDLcholesterol
in the body, the less uptake
of low-density lipoproteins (LDL) by tumor
cells which would suggest that there
is less lipid support for tumor growth75,
although additional work is needed to
understand this association more fully.
Statins, which are clinically used to lower
LDL levels in patients, have been considered
as a possible therapeutic target.
A recent study showed that treatment
with statins in VHL-deficient ccRCC
elicited promising early findings and
suggested that the observed lethality is
HIF dependent, highlighting statins as
promising therapeutic tools80.
Future Directions
Further analysis is needed for current treatments that can augment the current armamentarium for ccRCC. An area for growth in the research of therapeutic treatments is in targeting the metabolic dependencies, such as glycolysis, lipid, and cholesterol metabolism pathways, that discriminate ccRCC cells from normal tissues, or that reveal cellular adaptations associated with disease progression. In order to control glycogen metabolism in a favorable manner, promoting glycogen breakdown while simultaneously preventing glucose metabolism and glycogen synthesis is the goal. Glucagon is a natural substance in the body that accomplishes this by activating glycogen phosphorylase through the activity of protein kinase A. Finding a molecular target that can mimic this pathway specifically in ccRCC could be a direction worth pursuing. It is worth noting, glycogen breakdown to glucose- 1-phosphate feeds into glycolysis which could fuel growth, so another approach could involve a combination of nutrient restriction and current frontline therapies that impede cell growth and metabolism. There are no current studies that have examined the effects of dietary restrictions on ccRCC patients, but a correlation between BMI and the presence or absence of a VHL mutation in ccRCC patients has been observed81.
In considering lipid and cholesterol metabolism for therapeutic development, it is known how the inhibition of SCD leads to cholesterol accumulation, but there have been no further studies completed to show the relationship between VHL mutations and cholesterol synthesis. Secondly, while statins look to be a promising target and have shown to inhibit the proliferation of VHL-deficient ccRCC in vitro and in vivo, further analysis needs to be done on the efficacy, mode of action, and safety of these treatments. Also, since dietary lipid intake was shown to effect lipid buildup in the kidneys, further investigation should be conducted to determine outcomes when cholesterol treatments are compounded with dietary and host factors.
There is minimal literature in ccRCC investigating the role of acetate metabolism, an important branch of acetyl-CoA production and a key contributor to lipogenesis. Therefore, acetate metabolism and the enzyme acetate-dependent acetyl-CoA synthetase 2 (ACSS2) could be a potential therapeutic target. While this has not been explored in ccRCC, researchers have demonstrated in other tissues that inhibition of ACSS2 leads to the inhibition of lipid metabolism, changes to histone acetylation, and reduced tumor growth82. ACSS2 is required for acetate uptake and ACSS2 deficient mice were shown to have decreased liver tumor formation83. Nuclear ACSS2 synthesizes acetyl-CoA for histone acetylation, which activates lysozyme biogenesis84 Interestingly, it has been shown that acetyl-CoA derived from ACSS2 is required for the acetylation of HIF-2α and results in optimal signaling85. These factors make ACSS2 an enzyme of interest for further investigation. In summary, bioenergetic metabolism has long been recognized as a differentiating feature of ccRCC, and as we gain insights into these pathways and methods to intervene. Future work to incorporate these strategies in combination or in sequence with existing therapies will be a major opportunity to impact this metabolically driven disease.
KEYWORDS: Cytoreductive Partial Nephrectomy • Radical Nephrectomy • Renal Cell Carcinoma • Patient Selection
REFERENCES

Correspondence: W. Kimryn Rathmell, M.D., Ph.D., Vanderbilt University Medical Center Department of Medicine, 1161 21st Avenue South Suite D-3100, Medical Center North, Nashville, TN 37232. Phone: 615-343-8701; Fax: 616-343-2551; Email: Kimryn.Rathmell@ VUMC.org

Disclosures: None